TREEWEB
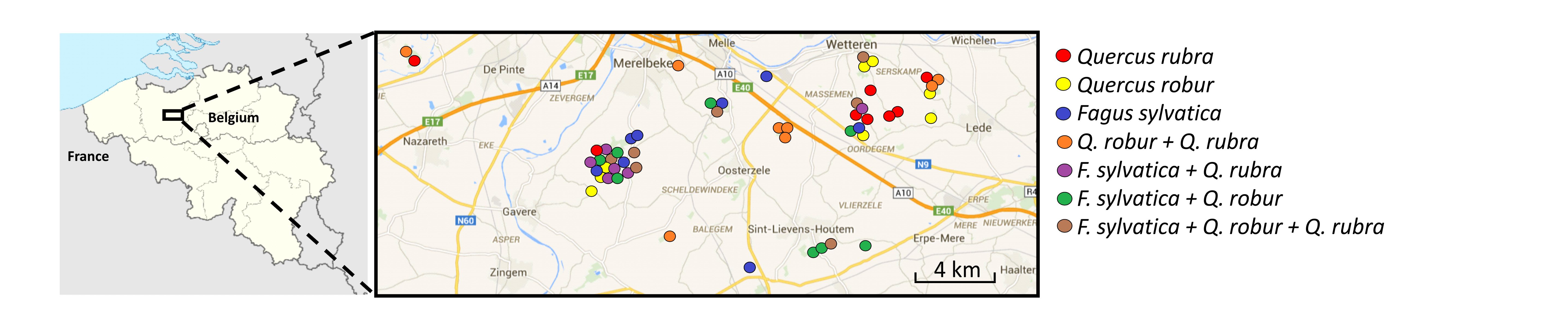
The acronym TREEWEB stands for the assessment of relationships between TREE diversity and ecosystem functioning across the terrestrial foodWEB, at different spatial scales. The project was launched in 2014 and will run until 2019. Interactions among detritivore, consumer and pathogen networks, which are tightly linked with forest primary production through feedback loops, will be studied in forests in East-Flanders, in a region demarcated by the cities of Gent, Deinze, Zottegem and Aalst. The forest plots lie in small or large ancient forest fragments and are either monocultures, 2-species mixtures or 3-species mixtures.
|
| The 53 TREEWEB plots |

|
| A beech plot with litter traps, and one of the birdboxes installed in the plot (October 2014) |
design
The plots are 30 m x 30 m large and are located in forest stands with a similar land-use history (ancient forest) and tree age (mature stands). The tree species pool includes the native Quercus robur and Fagus sylvatica and the non-native, locally invasive Q. rubra. For each of the seven tree species combinations (three monocultures, three 2-species mixtures, one 3-species mixture), four plots are set up in small forest fragments and four in large fragments. All tree species occur as monocultures and are represented at the two mixture levels, and a tree species is not present in each tree species combination. Thus, the design allows to distinguish tree diversity from tree identity effects. By implementing the design in large as well as small forest fragments, interactions between local variables, i.e., identity and diversity, and landscape features, i.e., connectivity, can also be studied.
research goals
Ultimately, the project aims to unravel how biological drivers operating at different spatial scales affect relationships between biodiversity and ecosystem functioning (photosynthesis, nutrient cycling, pest and disease dynamics). Tree diversity is predicted to affect forest ecosystem functioning through interactions among different subfoodwebs (detritivores, consumers, pathogens), which are tightly linked with forest primary production through feedback loops. To reach this goal we integrate (1) high-resolution sampling, measurement and observation schemes to infer correlative relationships, (2) strictly-controlled field and lab experiments to infer causal relationships, and (3) statistical modeling to infer generic relationships.
more info
For more info about this exploratory, send an e-mail to the contact person.